差异代谢物通路分析
KEGG(Kyoto Encyclopedia of Genes and Genomes)是一个整合了基因组、化学和系统功能信息的数据库[1],各个数据库中包含了大量的有用信息:基因组信息存储在GENES数据库里,包括完整和部分测序的基因组序列;更高级的功能信息存储在PATHWAY数据库里,包括图解的细胞生化过程如代谢、膜转运、信号传递、细胞周期,还包括同系保守的子通路等信息;KEGG的另一个数据 库是LIGAND,包含关于化学物质、酶分子、酶反应等信息。
KEGG提供的整合代谢途径(pathway)查询十分出色,包括碳水化合物、核苷、氨基酸等的代谢及有机物的生物降解,不仅提供了所有可能的代谢途径,而且对催化各步反应的酶进行了全面的注解,包括氨基酸序列、PDB库的链接等。KEGG是进行生物体内代谢分析、代谢网络研究的强有力工具,它有助于研究者把基因及其表达信息作为一个整体网络进行研究。
MetPA是metaboanalyst(www .metaboanalyst.ca)的一部分,主要基于KEGG代谢通路。MetPA数据库通过代谢通路浓缩和拓扑分析,识别出可能的受生物扰动的代谢通路,进而对代谢物的代谢通路进行分析。采用MetPA数据库可以分析两组差异代谢物的相关代谢通路,采用的数据分析算法是超几何检验,pathway拓扑结构采用的是Relative-betweeness Centrality。
基于上述MetPA分析结果,根据代谢通路中鉴定代谢物的相对响应值和降维算法,获得代谢通路的相对响应值,并以此计算代谢通路间的相关系数,绘制代谢通路关联网络图。
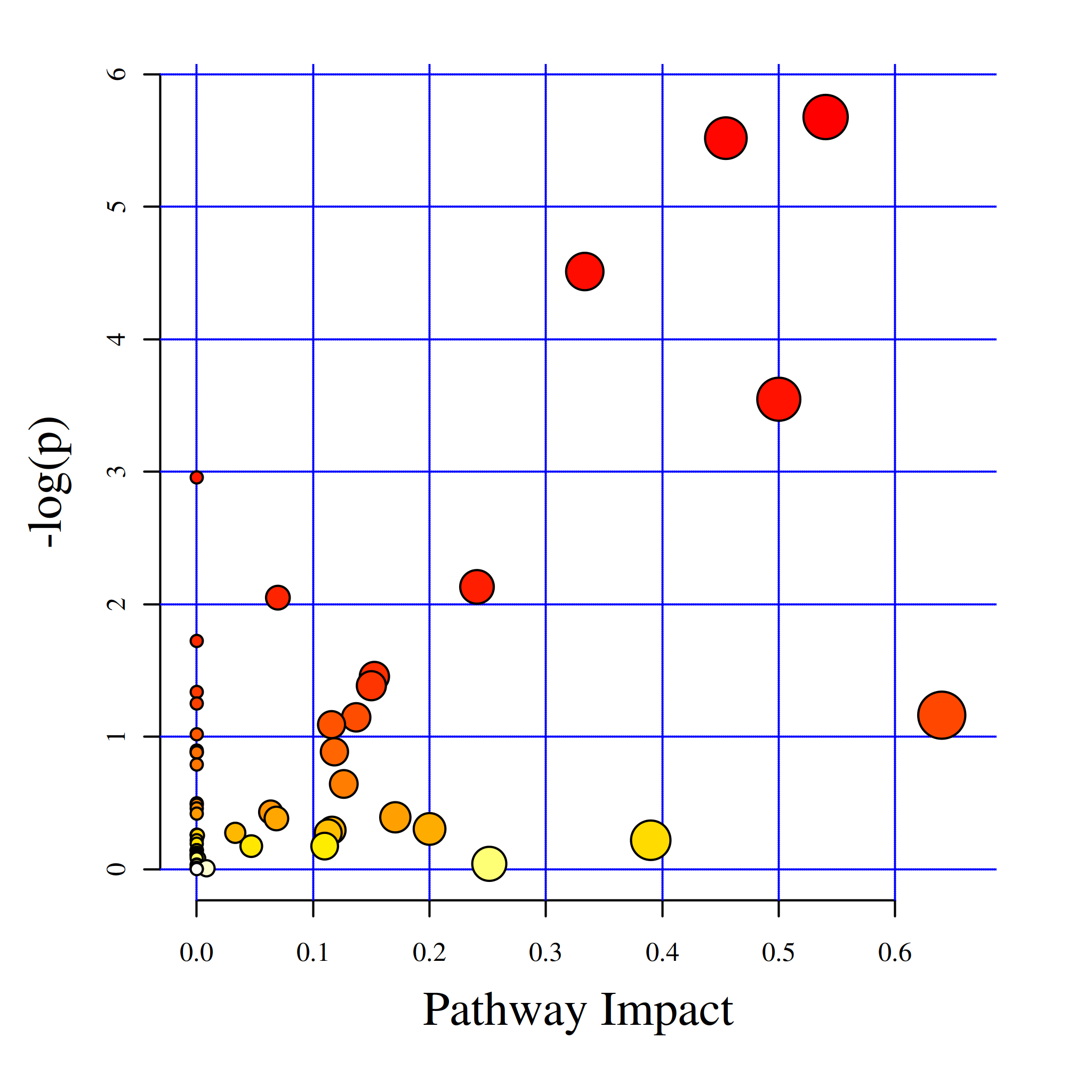
图:代谢通路影响因子气泡图
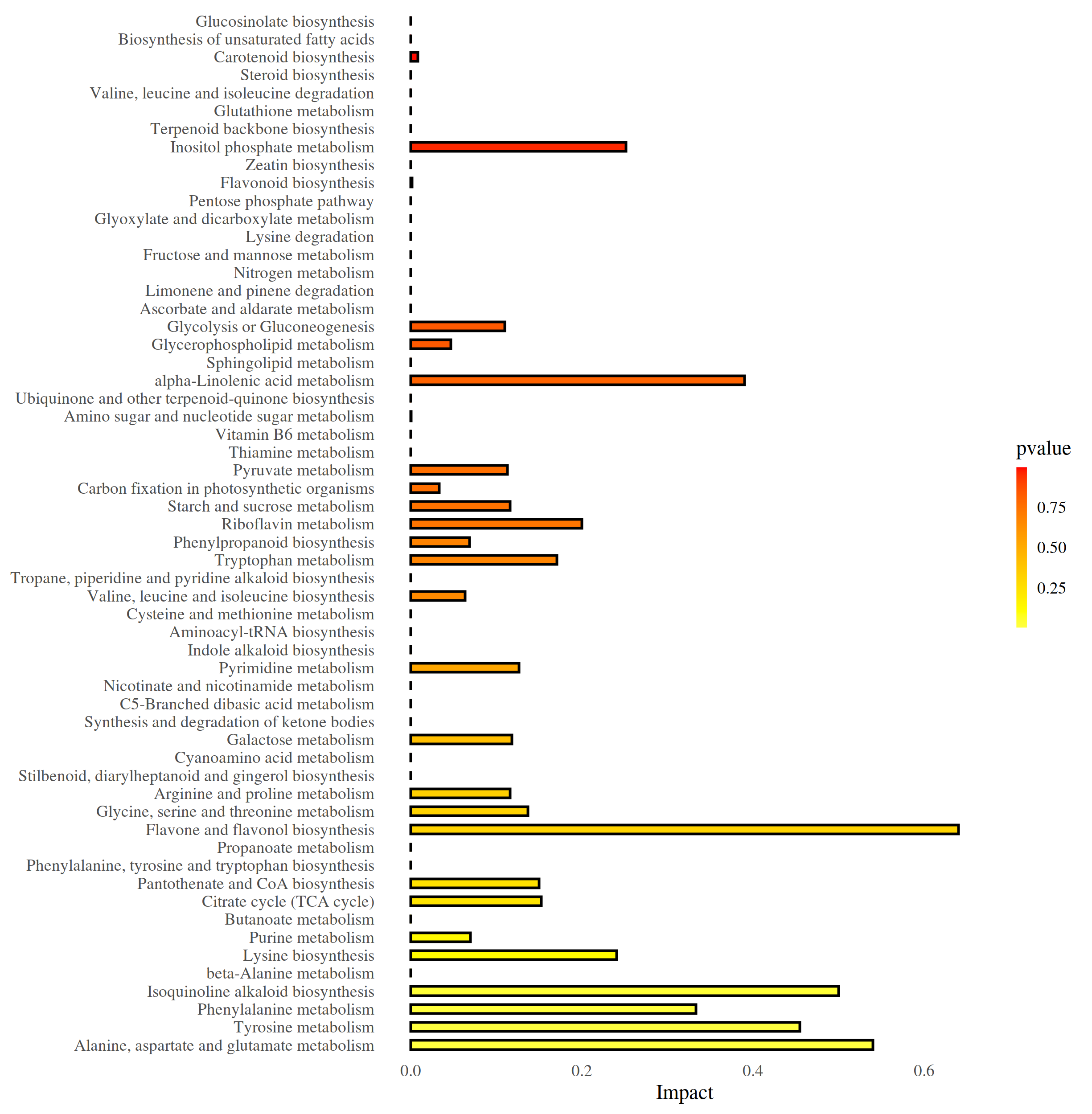
图:代谢通路影响因子柱状图
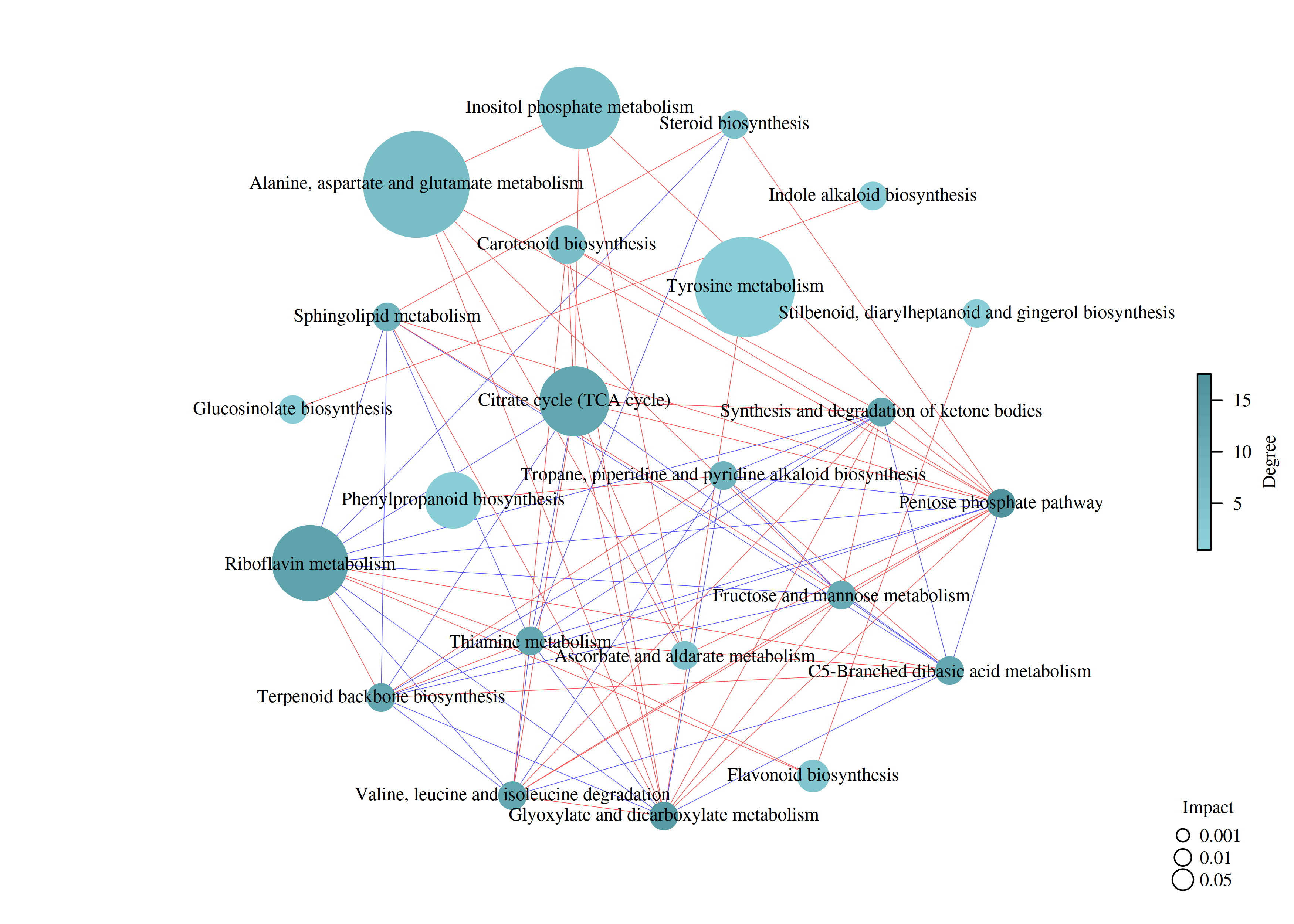
图:代谢通路网络图
| Total | Hits | Raw p | -LOG(p) | Holm adjust | FDR | Impact | compounds | pathway | |
|---|---|---|---|---|---|---|---|---|---|
| Alanine, aspartate and glutamate metabolism | 22 | 8 | 0.0034218 | 5.6776 | 0.29769 | 0.17459 | 0.54023 | C00049;C00026;C00064;C00152;C00122;C00022;C00042;C00334 | ath00250 |
| Tyrosine metabolism | 18 | 7 | 0.0040135 | 5.5181 | 0.34516 | 0.17459 | 0.45455 | C00355;C00082;C00483;C00122;C00164;C00042;C03765 | ath00350 |
| Phenylalanine metabolism | 8 | 4 | 0.010984 | 4.5113 | 0.93362 | 0.31853 | 0.33334 | C00166;C00423;C02505;C00811 | ath00360 |
| Isoquinoline alkaloid biosynthesis | 6 | 3 | 0.028801 | 3.5473 | 1 | 0.62642 | 0.5 | C00082;C00355;C00483 | ath00950 |
| beta-Alanine metabolism | 12 | 4 | 0.051969 | 2.9571 | 1 | 0.90426 | 0 | C00049;C00750;C00429;C00864 | ath00410 |
| Lysine biosynthesis | 10 | 3 | 0.11875 | 2.1308 | 1 | 1 | 0.24074 | C00666;C00049;C00047 | ath00300 |
| Purine metabolism | 61 | 11 | 0.12882 | 2.0493 | 1 | 1 | 0.06971 | C00064;C00212;C00559;C00262;C00242;C00366;C00362;C00147;C00294;C00387;C00330 | ath00230 |
| Butanoate metabolism | 18 | 4 | 0.17859 | 1.7227 | 1 | 1 | 0 | C00022;C00164;C00334;C00042 | ath00650 |
| Citrate cycle (TCA cycle) | 20 | 4 | 0.23357 | 1.4543 | 1 | 1 | 0.1526 | C00026;C00042;C00022;C00122 | ath00020 |
| Pantothenate and CoA biosynthesis | 14 | 3 | 0.2504 | 1.3847 | 1 | 1 | 0.15 | C00429;C00022;C00864 | ath00770 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 21 | 4 | 0.26247 | 1.3376 | 1 | 1 | 0 | C00463;C00166;C00082;C00078 | ath00400 |
| Propanoate metabolism | 15 | 3 | 0.28636 | 1.2505 | 1 | 1 | 0 | C00042;C05984;C00109 | ath00640 |
| Flavone and flavonol biosynthesis | 9 | 2 | 0.31278 | 1.1623 | 1 | 1 | 0.64 | C00389;C05623 | ath00944 |
| Glycine, serine and threonine metabolism | 30 | 5 | 0.31788 | 1.1461 | 1 | 1 | 0.13697 | C00049;C00188;C00109;C00022;C00078 | ath00260 |
| Arginine and proline metabolism | 38 | 6 | 0.33592 | 1.0909 | 1 | 1 | 0.1158 | C00049;C00062;C01250;C00064;C00122;C00334 | ath00330 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | 10 | 2 | 0.36129 | 1.0181 | 1 | 1 | 0 | C12208;C00852 | ath00945 |
| Cyanoamino acid metabolism | 11 | 2 | 0.40848 | 0.89531 | 1 | 1 | 0 | C00049;C06114 | ath00460 |
| Galactose metabolism | 26 | 4 | 0.41241 | 0.88573 | 1 | 1 | 0.11827 | C05401;C05399;C00267;C00137 | ath00052 |
| Synthesis and degradation of ketone bodies | 4 | 1 | 0.41421 | 0.88138 | 1 | 1 | 0 | C00164 | ath00072 |
| C5-Branched dibasic acid metabolism | 4 | 1 | 0.41421 | 0.88138 | 1 | 1 | 0 | C00022 | ath00660 |
| Nicotinate and nicotinamide metabolism | 12 | 2 | 0.45393 | 0.78981 | 1 | 1 | 0 | C00049;C05841 | ath00760 |
| Pyrimidine metabolism | 38 | 5 | 0.52567 | 0.64309 | 1 | 1 | 0.1263 | C00105;C00064;C00429;C00475;C00299 | ath00240 |
| Indole alkaloid biosynthesis | 7 | 1 | 0.60822 | 0.49722 | 1 | 1 | 0 | C00078 | ath00901 |
| Aminoacyl-tRNA biosynthesis | 67 | 8 | 0.61504 | 0.48607 | 1 | 1 | 0 | C00152;C00062;C00064;C00049;C00047;C00188;C00078;C00082 | ath00970 |
| Cysteine and methionine metabolism | 34 | 4 | 0.63202 | 0.45883 | 1 | 1 | 0 | C00491;C00049;C00022;C00109 | ath00270 |
| Valine, leucine and isoleucine biosynthesis | 26 | 3 | 0.65005 | 0.4307 | 1 | 1 | 0.06356 | C00188;C00022;C00109 | ath00290 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 8 | 1 | 0.65746 | 0.41937 | 1 | 1 | 0 | C00423 | ath00960 |
| Tryptophan metabolism | 27 | 3 | 0.67604 | 0.39151 | 1 | 1 | 0.17059 | C00078;C00632;C00780 | ath00380 |
| Phenylpropanoid biosynthesis | 45 | 5 | 0.68226 | 0.38234 | 1 | 1 | 0.06842 | C00811;C01494;C12208;C00423;C00852 | ath00940 |
| Riboflavin metabolism | 10 | 1 | 0.73824 | 0.30348 | 1 | 1 | 0.2 | C00255 | ath00740 |
| Starch and sucrose metabolism | 30 | 3 | 0.74543 | 0.2938 | 1 | 1 | 0.11621 | C00185;C00267;C00689 | ath00500 |
| Carbon fixation in photosynthetic organisms | 21 | 2 | 0.7602 | 0.27417 | 1 | 1 | 0.0331 | C00022;C00049 | ath00710 |
| Pyruvate metabolism | 21 | 2 | 0.7602 | 0.27417 | 1 | 1 | 0.11295 | C00022;C00227 | ath00620 |
| Thiamine metabolism | 11 | 1 | 0.77122 | 0.25979 | 1 | 1 | 0 | C04556 | ath00730 |
| Vitamin B6 metabolism | 11 | 1 | 0.77122 | 0.25979 | 1 | 1 | 0 | C00314 | ath00750 |
| Amino sugar and nucleotide sugar metabolism | 41 | 4 | 0.77533 | 0.25447 | 1 | 1 | 0.00057 | C00267;C00259;C01674;C00140 | ath00520 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 23 | 2 | 0.80391 | 0.21826 | 1 | 1 | 0 | C00082;C00811 | ath00130 |
| alpha-Linolenic acid metabolism | 23 | 2 | 0.80391 | 0.21826 | 1 | 1 | 0.39 | C04785;C06427 | ath00592 |
| Sphingolipid metabolism | 13 | 1 | 0.82529 | 0.19202 | 1 | 1 | 0 | C00836 | ath00600 |
| Glycerophospholipid metabolism | 25 | 2 | 0.84044 | 0.17383 | 1 | 1 | 0.04684 | C00588;C00670 | ath00564 |
| Glycolysis or Gluconeogenesis | 25 | 2 | 0.84044 | 0.17383 | 1 | 1 | 0.10989 | C00022;C00267 | ath00010 |
| Ascorbate and aldarate metabolism | 15 | 1 | 0.86664 | 0.14313 | 1 | 1 | 0 | C00137 | ath00053 |
| Limonene and pinene degradation | 15 | 1 | 0.86664 | 0.14313 | 1 | 1 | 0 | C11924 | ath00903 |
| Nitrogen metabolism | 15 | 1 | 0.86664 | 0.14313 | 1 | 1 | 0 | C00064 | ath00910 |
| Fructose and mannose metabolism | 16 | 1 | 0.88351 | 0.12386 | 1 | 1 | 0 | C00267 | ath00051 |
| Lysine degradation | 17 | 1 | 0.89825 | 0.1073 | 1 | 1 | 0 | C00047 | ath00310 |
| Glyoxylate and dicarboxylate metabolism | 17 | 1 | 0.89825 | 0.1073 | 1 | 1 | 0 | C00042 | ath00630 |
| Pentose phosphate pathway | 18 | 1 | 0.91114 | 0.093058 | 1 | 1 | 0 | C00257 | ath00030 |
| Flavonoid biosynthesis | 43 | 3 | 0.92146 | 0.081801 | 1 | 1 | 0.00137 | C12208;C00852;C00389 | ath00941 |
| Zeatin biosynthesis | 19 | 1 | 0.92241 | 0.08077 | 1 | 1 | 0 | C00147 | ath00908 |
| Inositol phosphate metabolism | 24 | 1 | 0.96067 | 0.040124 | 1 | 1 | 0.25131 | C00137 | ath00562 |
| Terpenoid backbone biosynthesis | 25 | 1 | 0.96568 | 0.034923 | 1 | 1 | 0 | C00022 | ath00900 |
| Glutathione metabolism | 26 | 1 | 0.97005 | 0.030403 | 1 | 1 | 0 | C01879 | ath00480 |
| Valine, leucine and isoleucine degradation | 34 | 1 | 0.98998 | 0.010068 | 1 | 1 | 0 | C00164 | ath00280 |
| Steroid biosynthesis | 36 | 1 | 0.99239 | 0.0076382 | 1 | 1 | 0 | C01802 | ath00100 |
| Carotenoid biosynthesis | 37 | 1 | 0.99337 | 0.0066527 | 1 | 1 | 0.0084 | C13455 | ath00906 |
| Biosynthesis of unsaturated fatty acids | 42 | 1 | 0.99667 | 0.0033313 | 1 | 1 | 0 | C06427 | ath01040 |
| Glucosinolate biosynthesis | 54 | 1 | 0.99937 | 0.00062737 | 1 | 1 | 0 | C00078 | ath00966 |
注:Total,目标代谢通路中代谢物的总数;Hits,目标代谢通路中差异代谢物数量,Raw p,超几何分布检验的p值;-log(p):对p值的自然对数取负值;Holm adjust,Holm假阳性矫正后p值;FDR,假阳性校正后值;Impact,代谢通路影响值;compounds,代谢物KEGG ID;pathway,代谢物代谢通路ID








