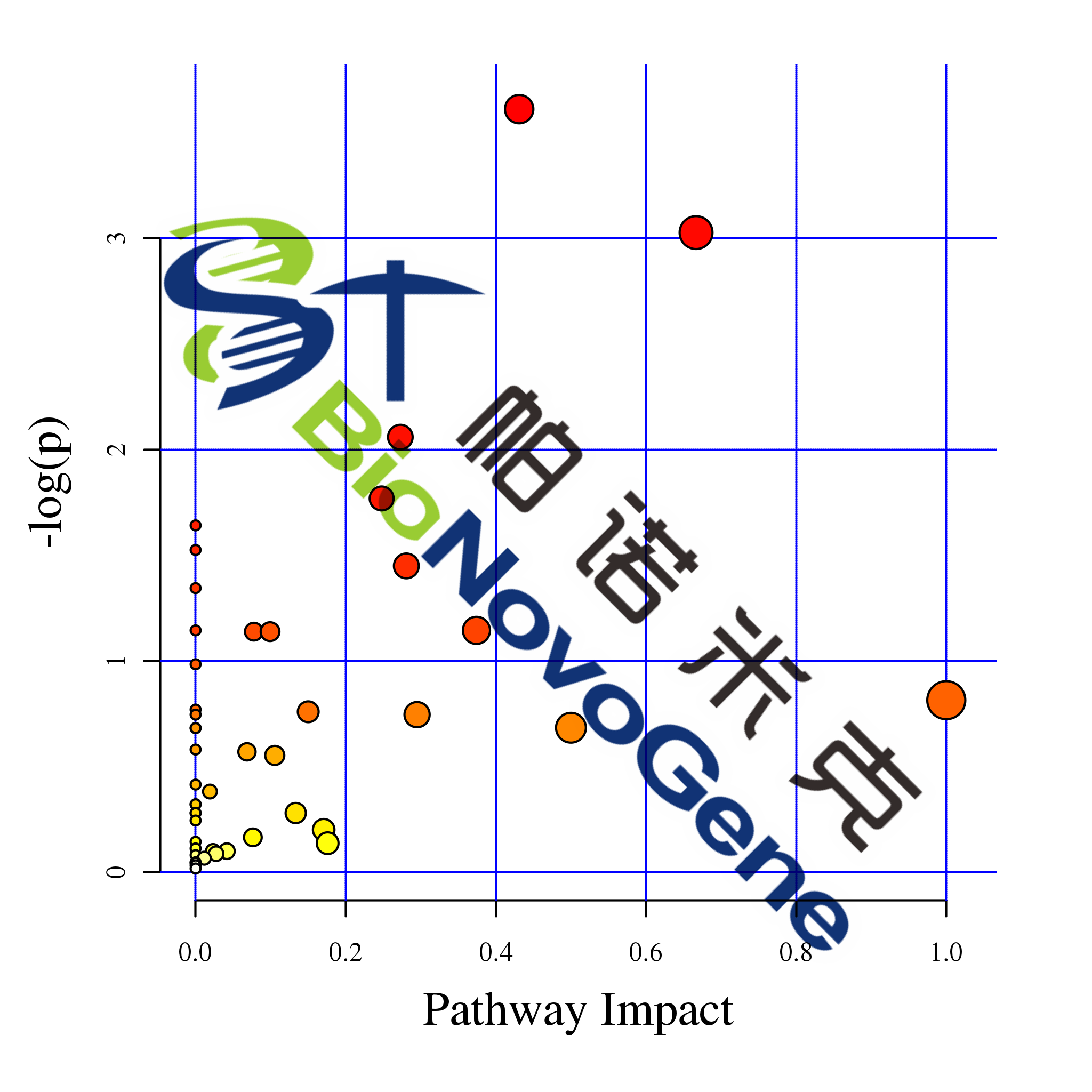
差异代谢物通路分析
KEGG(Kyoto Encyclopedia of Genes and Genomes)是一个整合了基因组、化学和系统功能信息的数据库[1],各个数据库中包含了大量的有用信息:基因组信息存储在GENES数据库里,包括完整和部分测序的基因组序列;更高级的功能信息存储在PATHWAY数据库里,包括图解的细胞生化过程如代谢、膜转运、信号传递、细胞周期,还包括同系保守的子通路等信息;KEGG的另一个数据 库是LIGAND,包含关于化学物质、酶分子、酶反应等信息。
KEGG提供的整合代谢途径(pathway)查询十分出色,包括碳水化合物、核苷、氨基酸等的代谢及有机物的生物降解,不仅提供了所有可能的代谢途径,而且对催化各步反应的酶进行了全面的注解,包括氨基酸序列、PDB库的链接等。KEGG是进行生物体内代谢分析、代谢网络研究的强有力工具,它有助于研究者把基因及其表达信息作为一个整体网络进行研究。
MetPA是metaboanalyst(www .metaboanalyst.ca)的一部分,主要基于KEGG代谢通路。MetPA数据库通过代谢通路浓缩和拓扑分析,识别出可能的受生物扰动的代谢通路,进而对代谢物的代谢通路进行分析。采用MetPA数据库可以分析两组差异代谢物的相关代谢通路,采用的数据分析算法是超几何检验,pathway拓扑结构采用的是Relative-betweeness Centrality。
图:代谢通路影响因子图
DOWNLOAD
| Total | Hits | Raw p | -LOG(p) | Holm adjust | FDR | Impact | compounds | pathway | |
|---|---|---|---|---|---|---|---|---|---|
| Alanine, aspartate and glutamate metabolism | 22 | 5 | 0.10504 | 2.2534 | 1 | 1 | 0.42528 | C00232;C00026;C00025;C00152;C00334 | ath00250 |
| Isoquinoline alkaloid biosynthesis | 6 | 2 | 0.14935 | 1.9015 | 1 | 1 | 0 | C00355;C00483 | ath00950 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 8 | 2 | 0.23921 | 1.4304 | 1 | 1 | 0 | C00079;C00423 | ath00960 |
| Phenylalanine metabolism | 8 | 2 | 0.23921 | 1.4304 | 1 | 1 | 0.66667 | C00079;C00423 | ath00360 |
| Aminoacyl-tRNA biosynthesis | 67 | 10 | 0.25208 | 1.378 | 1 | 1 | 0 | C00152;C00079;C00062;C00097;C00183;C00047;C00123;C00078;C00148;C00025 | ath00970 |
| Flavone and flavonol biosynthesis | 9 | 2 | 0.28528 | 1.2543 | 1 | 1 | 0 | C10098;C12249 | ath00944 |
| Butanoate metabolism | 18 | 3 | 0.3547 | 1.0365 | 1 | 1 | 0 | C00025;C00232;C00334 | ath00650 |
| Tyrosine metabolism | 18 | 3 | 0.3547 | 1.0365 | 1 | 1 | 0.18182 | C00355;C00232;C00483 | ath00350 |
| Pentose phosphate pathway | 18 | 3 | 0.3547 | 1.0365 | 1 | 1 | 0.32269 | C00117;C05382;C00257 | ath00030 |
| Carbon fixation in photosynthetic organisms | 21 | 3 | 0.45455 | 0.78845 | 1 | 1 | 0.03644 | C00149;C05382;C00117 | ath00710 |
| Arginine and proline metabolism | 38 | 5 | 0.4657 | 0.76421 | 1 | 1 | 0.36738 | C00327;C00062;C00025;C00148;C00334 | ath00330 |
| Pantothenate and CoA biosynthesis | 14 | 2 | 0.502 | 0.68916 | 1 | 1 | 0.15 | C00183;C00864 | ath00770 |
| Nitrogen metabolism | 15 | 2 | 0.54028 | 0.61567 | 1 | 1 | 0 | C00079;C00025 | ath00910 |
| Fructose and mannose metabolism | 16 | 2 | 0.57649 | 0.55079 | 1 | 1 | 0.07627 | C00794;C00095 | ath00051 |
| Indole alkaloid biosynthesis | 7 | 1 | 0.58316 | 0.53929 | 1 | 1 | 0 | C00078 | ath00901 |
| Glutathione metabolism | 26 | 3 | 0.60419 | 0.50387 | 1 | 1 | 0.07756 | C01879;C00025;C00097 | ath00480 |
| Galactose metabolism | 26 | 3 | 0.60419 | 0.50387 | 1 | 1 | 0.3457 | C00124;C00095;C00794 | ath00052 |
| Phenylpropanoid biosynthesis | 45 | 5 | 0.6233 | 0.47273 | 1 | 1 | 0.05192 | C01494;C00079;C00423;C00852;C00903 | ath00940 |
| Tryptophan metabolism | 27 | 3 | 0.63071 | 0.4609 | 1 | 1 | 0.37647 | C00078;C00632;C02938 | ath00380 |
| Citrate cycle (TCA cycle) | 20 | 2 | 0.70033 | 0.3562 | 1 | 1 | 0.10521 | C00026;C00149 | ath00020 |
| Caffeine metabolism | 10 | 1 | 0.71396 | 0.33692 | 1 | 1 | 0 | C13747 | ath00232 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | 10 | 1 | 0.71396 | 0.33692 | 1 | 1 | 0 | C00852 | ath00945 |
| Lysine biosynthesis | 10 | 1 | 0.71396 | 0.33692 | 1 | 1 | 0.07407 | C00047 | ath00300 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 21 | 2 | 0.7262 | 0.31992 | 1 | 1 | 0 | C00078;C00079 | ath00400 |
| Thiamine metabolism | 11 | 1 | 0.74776 | 0.29067 | 1 | 1 | 0 | C00097 | ath00730 |
| Vitamin B6 metabolism | 11 | 1 | 0.74776 | 0.29067 | 1 | 1 | 0 | C00314 | ath00750 |
| Flavonoid biosynthesis | 43 | 4 | 0.76284 | 0.27071 | 1 | 1 | 0.17584 | C00509;C02906;C01617;C00852 | ath00941 |
| beta-Alanine metabolism | 12 | 1 | 0.77759 | 0.25156 | 1 | 1 | 0 | C00864 | ath00410 |
| Sulfur metabolism | 12 | 1 | 0.77759 | 0.25156 | 1 | 1 | 0.13333 | C00097 | ath00920 |
| Fatty acid elongation in mitochondria | 13 | 1 | 0.80391 | 0.21827 | 1 | 1 | 0 | C00249 | ath00062 |
| Valine, leucine and isoleucine biosynthesis | 26 | 2 | 0.82886 | 0.1877 | 1 | 1 | 0.01865 | C00123;C00183 | ath00290 |
| Fatty acid biosynthesis | 49 | 4 | 0.84663 | 0.16649 | 1 | 1 | 0 | C08362;C01571;C02679;C00249 | ath00061 |
| Lysine degradation | 17 | 1 | 0.88164 | 0.12597 | 1 | 1 | 0 | C00047 | ath00310 |
| Glyoxylate and dicarboxylate metabolism | 17 | 1 | 0.88164 | 0.12597 | 1 | 1 | 0.15986 | C00149 | ath00630 |
| Glycine, serine and threonine metabolism | 30 | 2 | 0.88462 | 0.12259 | 1 | 1 | 0 | C00719;C00078 | ath00260 |
| Glucosinolate biosynthesis | 54 | 4 | 0.89612 | 0.10968 | 1 | 1 | 0 | C00079;C00078;C00183;C00123 | ath00966 |
| Valine, leucine and isoleucine degradation | 34 | 2 | 0.92323 | 0.079878 | 1 | 1 | 0 | C00183;C00123 | ath00280 |
| Pyruvate metabolism | 21 | 1 | 0.92868 | 0.073993 | 1 | 1 | 0.08561 | C00149 | ath00620 |
| alpha-Linolenic acid metabolism | 23 | 1 | 0.94467 | 0.056919 | 1 | 1 | 0 | C11512 | ath00592 |
| Porphyrin and chlorophyll metabolism | 29 | 1 | 0.97424 | 0.026102 | 1 | 1 | 0 | C00025 | ath00860 |
| Fatty acid metabolism | 34 | 1 | 0.98641 | 0.01368 | 1 | 1 | 0 | C00249 | ath00071 |
| Cysteine and methionine metabolism | 34 | 1 | 0.98641 | 0.01368 | 1 | 1 | 0.04167 | C00097 | ath00270 |
| Carotenoid biosynthesis | 37 | 1 | 0.99076 | 0.009286 | 1 | 1 | 0.0056 | C06082 | ath00906 |
| Pyrimidine metabolism | 38 | 1 | 0.99187 | 0.0081605 | 1 | 1 | 0.00104 | C00526 | ath00240 |
| Amino sugar and nucleotide sugar metabolism | 41 | 1 | 0.99448 | 0.0055369 | 1 | 1 | 0 | C00124 | ath00520 |
| Biosynthesis of unsaturated fatty acids | 42 | 1 | 0.99515 | 0.0048648 | 1 | 1 | 0 | C00249 | ath01040 |
| Purine metabolism | 61 | 2 | 0.99609 | 0.0039147 | 1 | 1 | 0.01472 | C00117;C00362 | ath00230 |
注:Total,目标代谢通路中代谢物的总数;Hits,目标代谢通路中差异代谢物数量,Raw p,超几何分布检验的p值;-log(p):对p值的自然对数取负值;Holm adjust,Holm假阳性矫正后p值;FDR,假阳性校正后值;Impact,代谢通路影响值;compounds,代谢物KEGG ID;pathway,代谢物代谢通路ID